Optimisation of Sensor Integration in Organ-on-Chip Platforms
Organs-on-Chips (OoCs) are microfluidics-based systems that aim to recapitulate the in-vivo behaviour of specific organs. Using a network of microfluidic channels, OoCs provide a biophysical and biochemical environment to sustain complex 2D and 3D cell cultures systems that recapitulate physiological conditions. Such systems are expected to reduce the need for animal testing, accelerate drug discovery and improve the prediction of drug efficacy and toxicity.
OoCs can be interfaced with biosensors for real-time and high-throughput measurements. However, such an integration comes with its own challenges. First, the timescales in biosensing or OoCs are significantly different. Biosensors are typically optimised to provide near-real time answers; however, they can rarely operate continuously without degradation of performance. In contrast, OoC operation can last several days or weeks. In addition, due to the limited number of cells, changes in molecular concentrations in OoCs can be extremely low. Finally, cells in culture may be affected by shear stress, which limits the flow rate for a given chamber dimension.
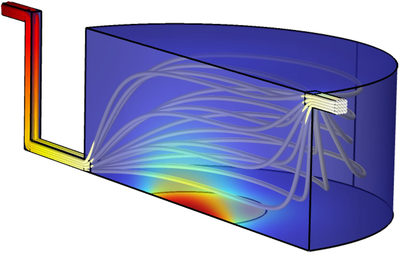
To address these complex challenges, we use the COMSOL Multiphysics® software with the Laminar Flow, Transport of Diluted Species and General Form Boundary PDE physics interfaces for the optimisation of the flow conditions and dimensions of the platform. The parametric model consists of a cell culture chamber connected to an inlet to modulate the flow and an outlet where the sensor is located (Fig. 1). A multimodal sensor is used to monitor specific cellular cytokine/chemokine production and glucose consumption. Due to cytotoxic production of H2O2 generated by the amperometric glucose sensors, it is placed downstream from the culture chamber.
We used experimental values in static mode, to define the flux of the molecules of interest. Using a first model, we demonstrated that an incubation time (without flow) was necessary to accumulate enough molecular concentration changes in the chamber before transferring its content to the sensor. Indeed, a continuous flow dilutes the chamber content and prevents reaching the sensor working range. To optimise the simulation, a two-step study to account for the incubation (hours timescale) and the active transport (minutes timescale) was set-up. A parametric sweep in the second study enables the evaluation of several flow and molecular conditions. Incubation time and chamber geometry were also studied systematically.
Figure 2a and b show the normalised concentration at the sensor surface for two geometries. In each case, the incubation time is 1h (3600 s) and the injection time 10 minutes (600 s) and the flow rates vary between 1 to 100 mL/h. The figures show that the concentration at the sensor depends on the flow rate. Indeed, a slow flow rate does not bring enough molecules to the sensors surface in the given timeframe, whereas high flow rate results in a liquid dilution. It is noted that these values are not absolute and depend on the volume of the chamber. In all our simulations, the flow rate was upper bound by a pre-defined shear stress value to minimise cells damage. A new model accounts for molecular adsorption at the sensor’s interface.
Herunterladen
- Charmet_4821_poster.pdf - 2.63MB
