Simulating Organogenesis in COMSOL Multiphysics®: Cell-based Signaling Models
Veröffentlicht in 2013
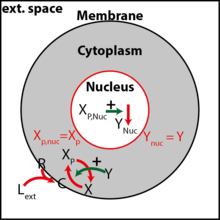
Most models of biological pattern formation are simulated on continuous domains even though cells are discrete objects that provide internal boundaries to the diffusion of regulatory components. In our previous papers on simulating organogenesis in COMSOL Multiphysics® (Germann et al COMSOL Multiphysics® Conf Procedings 2011; Menshykau and Iber, COMSOL Multiphysics® Conf Proceedings 2012) we discussed methods to efficiently solve signalling models on static and growing continuous domains. Here we discuss COMSOL Multiphysics®-based methods to study spatio-temporal signaling models in a tissue at cellular resolution with subcellular compartments, i.e. cell membrane, cytoplasm, and nucleus.
Herunterladen
- iber_poster.pdf - 0.59MB
- iber_paper.pdf - 0.91MB
- iber_abstract.pdf - 0.02MB



